OUR PRODUCTS
Products that make metabolomics easier
WORKFLOW KIT
MASS SPECTROMETRY
METABOLIC PROFILING
OUR TECHNOLOGY
Learn how we use internal standards as metabolic profiling tools to eliminate noise, analytical variability and ion suppression. Internal Standards make it easier to achieve clean, accurate and reproducible datasets. Better data translates into better results.
C13-labelled Yeast Metabolomics Workflow

How IROA Works

IROA MLSDiscovery Software

Metabolomics Compound Identification
PUBLICATIONS
Read our publications on metabolic profiling technologies that automate the measurement of biochemicals in biological systems.
Metabolomics Research
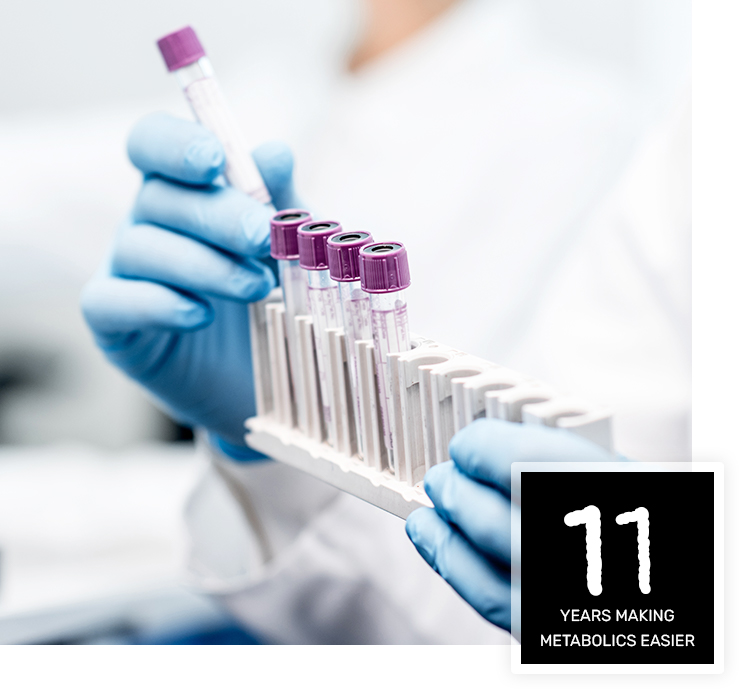
ABOUT US
Metabolomics Company founded in 2010,
Ann Arbor, Michigan.
Our mission is to simply metabolomics measurements. Uniquely-labeled internal standards are key to our technology because they are easily identified in mass spectral data using software algorithms. When spiked into biological samples, the internal standards allow the quantitation of 100s of metabolites in blood, urine, biopsies etc.
Listen To Webinar
The IROA TruQuant IQQ Cannabis metabolic profiling workflow provides a software solution together with a carefully formulated Cannabis Internal Standard for accurate simultaneous measurement of 100s of compounds.
Some of our Customers
Metric

NC State University

Senda

Stanford Health Care School of Medicine